In this article, we will explore the process of visualizing K-means clustering results using two popular R packages: factoextra and ggpubr.
Required R Packages
To follow along with this guide, you'll need to install and load the following R packages:
ggpubrfactoextra
You can install these packages by running the following code:
library(ggpubr)
library(factoextra)
Data Preparation
For this example, we will use the famous iris dataset. This dataset contains 150 samples from three species of iris (setosa, versicolor, and virginica). Each sample is described by four variables: sepal length, sepal width, petal length, and petal width.
data("iris")
df <- iris
head(df, 3)
K-Means Clustering Calculation Example
To perform K-means clustering on the iris dataset, we will remove the species column (the fifth column) and scale the remaining variables using the scale() function. Then, we'll use the kmeans() function from the stats package to cluster the data.
# Scale the data
df_scaled <- scale(df[, -5])
# Perform K-means clustering
set.seed(123)
kmeans_result <- kmeans(df_scaled, centers = 3)
# Print the cluster assignments
print(kmeans_result$cluster)
Visualizing K-Means Clusters with factoextra
To visualize the K-means clusters using factoextra, we'll first compute the individual coordinates of each sample using principal component analysis (PCA). Then, we'll add the cluster assignments and species labels to the coordinate data.
# Compute PCA
res.pca <- prcomp(df_scaled, scale = TRUE)
# Extract individual coordinates
ind.coord <- as.data.frame(get_pca_ind(res.pca)$coord)
# Add cluster assignments and species labels
ind.coord$cluster <- factor(kmeans_result$cluster)
ind.coord$Species <- df$Species
# Data inspection
head(ind.coord)
Visualizing K-Means Clusters with ggpubr
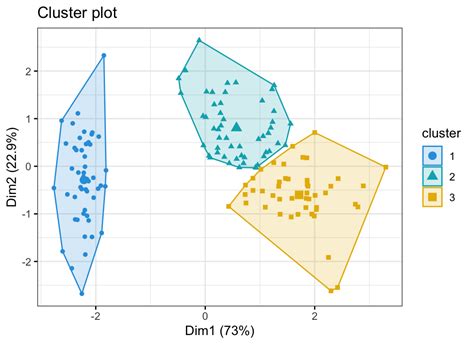
To visualize the K-means clusters using ggpubr, we'll create a scatter plot of the first two PCA dimensions, color-coded by cluster assignments. We'll also add concentration ellipses and cluster centroids to the plot.
# Create the scatter plot
ggscatter(
ind.coord, x = "Dim.1", y = "Dim.2",
color = "cluster", palette = "npg", ellipse = TRUE, ellipse.type = "convex",
shape = "Species", size = 1.5, legend = "right", ggtheme = theme_bw(),
xlab = paste0("Dim 1 (", variance.percent[1], "% )" ),
ylab = paste0("Dim 2 (", variance.percent[2], "% )" )
) +
stat_mean(aes(color = cluster), size = 4)
This plot shows the K-means clusters in a two-dimensional representation, with each point colored according to its cluster assignment and labeled by species. The concentration ellipses represent the distribution of points within each cluster.
###In this article, we've demonstrated how to visualize K-means clustering results using factoextra and ggpubr in R. By following these step-by-step examples, you can easily create interactive visualizations of your own K-means clustering results.